Tutorial 10 Short Read Alignment Part 3 Igv Visualization

Igv Screenshot Alignment Of Wt Sv02 B C Igv Screenshot Of The tutorials are organized into clear and progressive modules. 📺in this bioinformatics tutorial whether you're new to bioinformatics or looking to enhance your skills in genomics and. For this part, we will be using publicly available illumina sequence data generated for the hcc1143 cell line. the hcc1143 cell line was generated from a 52 year old caucasian woman with breast cancer.
Igv Browser V2 4 View Of Read Alignment Of The Largest Rearrangement Educational tutorials and working pipelines for rna seq analysis including an introduction to: cloud computing, critical file formats, reference genomes, gene annotation, expression, differential expression, alternative splicing, data visualization, and interpretation. Desktop application for the interactive visual exploration of integrated genomic datasets. integrative genomics viewer (igv) with igv you can… • explore large genomic datasets with an intuitive, easy to use interface. • integrate multiple data types with clinical and other sample information. More recently we have developed a suite of specialized features to support third generation long read sequencing technologies. here we present a short overview of igv variant visualization capabilities illustrated with examples drawn from cancer and germline datasets. Read mapping is aligning each of your reads to a the matching locus on a longer sequence (assembly). you could think of it at the location of where your read would hybridize to your genome if you could do this experiment. usually few mismatches are allowed (think about the consequences).

Igv Browser V2 4 View Of Read Alignment Of The Largest Rearrangement More recently we have developed a suite of specialized features to support third generation long read sequencing technologies. here we present a short overview of igv variant visualization capabilities illustrated with examples drawn from cancer and germline datasets. Read mapping is aligning each of your reads to a the matching locus on a longer sequence (assembly). you could think of it at the location of where your read would hybridize to your genome if you could do this experiment. usually few mismatches are allowed (think about the consequences). Visualizing the results with igv (web based or desktop) or tablet. provide clear instructions with examples. include explanations of each step and the purpose of the tools used. ensure the reference genome is indexed before alignment. use a publicly available wgs seq dataset from geo: gse186573. crc: colorectal cancer. stad: stomach adenocarcinoma. We will be visualizing read alignments using igv, a popular visualization tool for hts data. first, lets familiarize ourselves with it. by default, igv loads the human grch38 hg38 reference genome. While that is running, let’s take a look at an alignment in igv. we are going to take an already done alignment and cut it down so that it is small enough to download easily. Visualizing the results with igv (web based or desktop) or tablet. provide clear instructions with examples. include explanations of each step and the purpose of the tools used. input file: human genome data (subset to keep it manageable). reference genome: grch38 (human genome assembly).
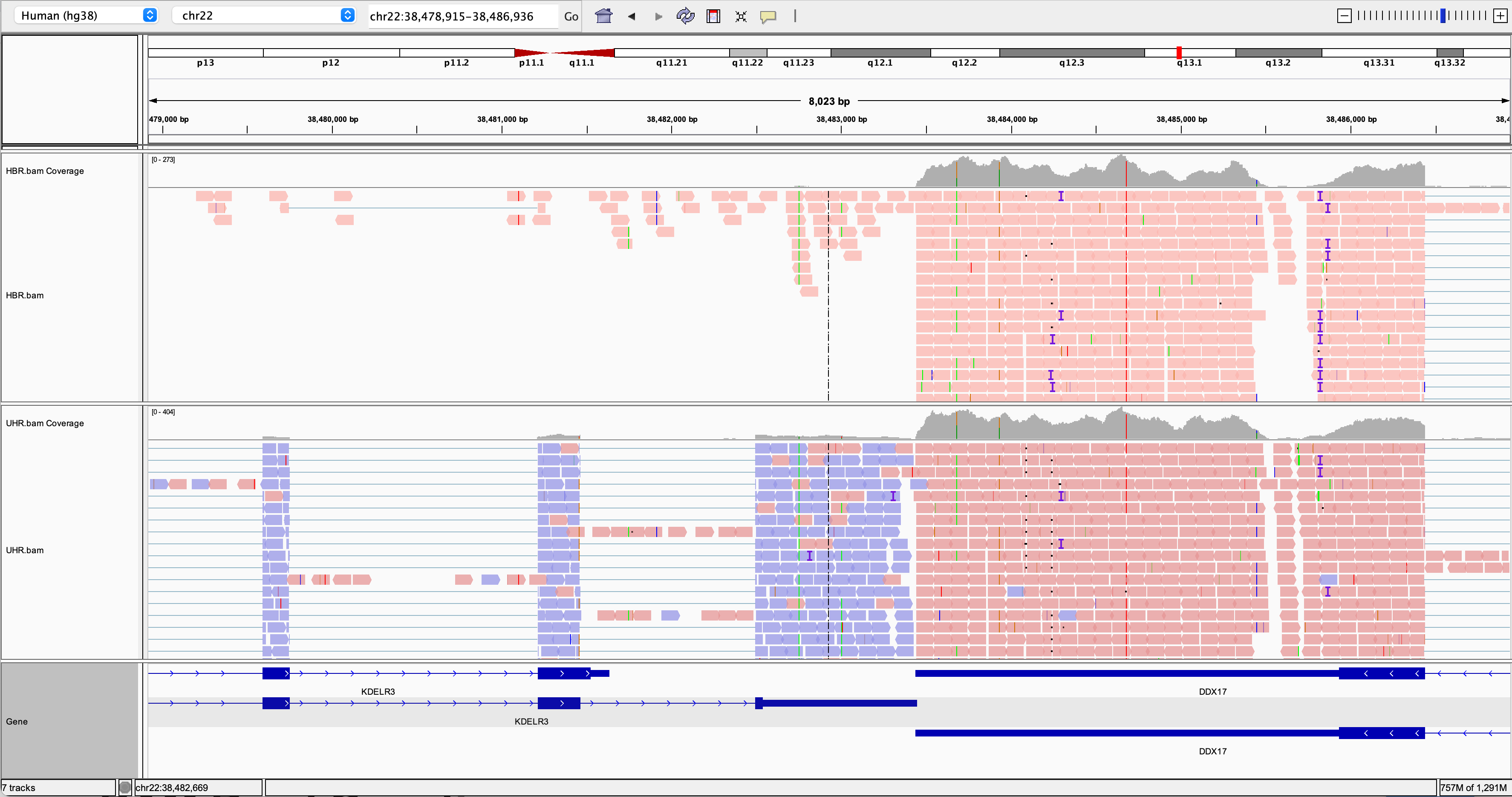
Alignment Visualization Igv Griffith Lab Visualizing the results with igv (web based or desktop) or tablet. provide clear instructions with examples. include explanations of each step and the purpose of the tools used. ensure the reference genome is indexed before alignment. use a publicly available wgs seq dataset from geo: gse186573. crc: colorectal cancer. stad: stomach adenocarcinoma. We will be visualizing read alignments using igv, a popular visualization tool for hts data. first, lets familiarize ourselves with it. by default, igv loads the human grch38 hg38 reference genome. While that is running, let’s take a look at an alignment in igv. we are going to take an already done alignment and cut it down so that it is small enough to download easily. Visualizing the results with igv (web based or desktop) or tablet. provide clear instructions with examples. include explanations of each step and the purpose of the tools used. input file: human genome data (subset to keep it manageable). reference genome: grch38 (human genome assembly).

Visualization Of Read Alignments Image Produced With A Modified While that is running, let’s take a look at an alignment in igv. we are going to take an already done alignment and cut it down so that it is small enough to download easily. Visualizing the results with igv (web based or desktop) or tablet. provide clear instructions with examples. include explanations of each step and the purpose of the tools used. input file: human genome data (subset to keep it manageable). reference genome: grch38 (human genome assembly).
Comments are closed.