Methods For Cell Type Deconvolution And Estimation Of Tumour Purity

Methods For Cell Type Deconvolution And Estimation Of Tumour Purity We discuss several computational tools, which enable deconvolution of genomic and transcriptomic data from heterogeneous samples. we also performed a systematic comparative assessment of these tools. We present methods to refine data simulation with real tumor data guidance to generate a modest training set and tailor a unified deep learning model to bypass the issue of highly variable gene expression in cancer cells.

Methods For Cell Type Deconvolution And Estimation Of Tumour Purity Variation in tumor purity (proportion of cancer cells in a sample) can both confound integrative analysis and enable studies of tumor heterogeneity. here we developed puree, which uses a. Other reference free and semi referencefree approaches employ methods such as nmf and recursive quantile projection (qp) which have been used to estimate cell type proportions. Co quantified transcripts and proteins performed similarly to estimate stroma and immune cell admixture (r ≥ 0.63) in two commonly used deconvolution algorithms, estimate or consensus tme. Here, we present an overview of 20 deconvolution techniques, including cutting edge techniques recently established. we categorized deconvolution techniques by three primary criteria: characteristics of methodology, use of prior knowledge of cell types and outcome of the methods.

Cell Type Deconvolution Analyses Of Tumour Cell Specific Clusters From Co quantified transcripts and proteins performed similarly to estimate stroma and immune cell admixture (r ≥ 0.63) in two commonly used deconvolution algorithms, estimate or consensus tme. Here, we present an overview of 20 deconvolution techniques, including cutting edge techniques recently established. we categorized deconvolution techniques by three primary criteria: characteristics of methodology, use of prior knowledge of cell types and outcome of the methods. Here, we propose methylbert, a transformer based model for read level methylation pattern classification. methylbert identifies tumour derived sequence reads based on their methylation patterns. Because tumor purity is a crucial factor in cancer research, various methods have been developed to estimate it, including dna 1 , dna methylation 2 , and rna 3,4 based deconvolution approaches. Deconbench performs a systematic performance evaluation of each new methodological contribution and provides the possibility to publically share source code and scoring. Infiltrating stromal and immune cells form the major fraction of normal cells in tumour tissue and not only perturb the tumour signal in molecular studies but also have an important role in.

Tumour Deconvolution And The Estimation Adjustment Results For Dlbcl Here, we propose methylbert, a transformer based model for read level methylation pattern classification. methylbert identifies tumour derived sequence reads based on their methylation patterns. Because tumor purity is a crucial factor in cancer research, various methods have been developed to estimate it, including dna 1 , dna methylation 2 , and rna 3,4 based deconvolution approaches. Deconbench performs a systematic performance evaluation of each new methodological contribution and provides the possibility to publically share source code and scoring. Infiltrating stromal and immune cells form the major fraction of normal cells in tumour tissue and not only perturb the tumour signal in molecular studies but also have an important role in.
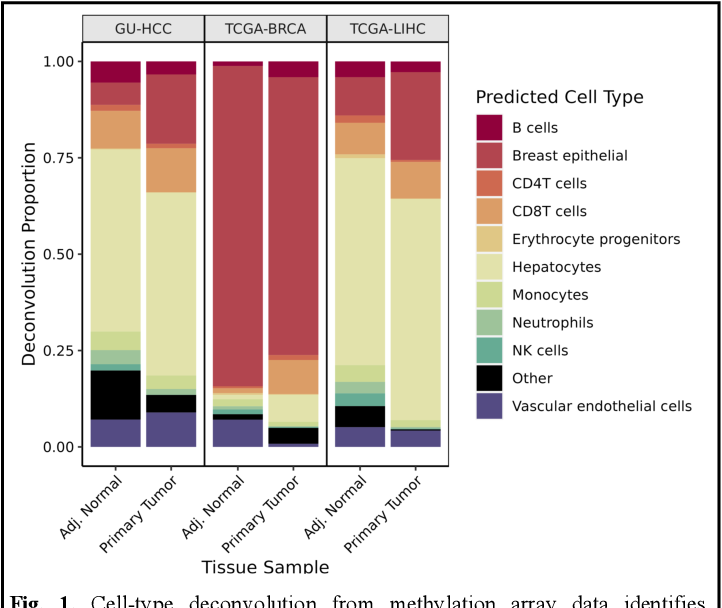
Figure 1 From Cell Type Deconvolution And Age Estimation Using Dna Deconbench performs a systematic performance evaluation of each new methodological contribution and provides the possibility to publically share source code and scoring. Infiltrating stromal and immune cells form the major fraction of normal cells in tumour tissue and not only perturb the tumour signal in molecular studies but also have an important role in.
Comments are closed.