Introduction And Workflow Of Single Cell Rna Seq Part1 Bioinformatics Course

Pdf Introduction Into Single Cell Rna Seq Ut Single Cell Rna In this video, i'll guide you through the basics of single cell rna seq analysis and the workflow. learn how single cell analysis is transforming our understanding of biology by. In contrast to drop seq, where solid beads are used for rna capture, 10x uses soft hydrogels containing oligos. these enable “single poisson loading” leading to capture of >60% of input cells.
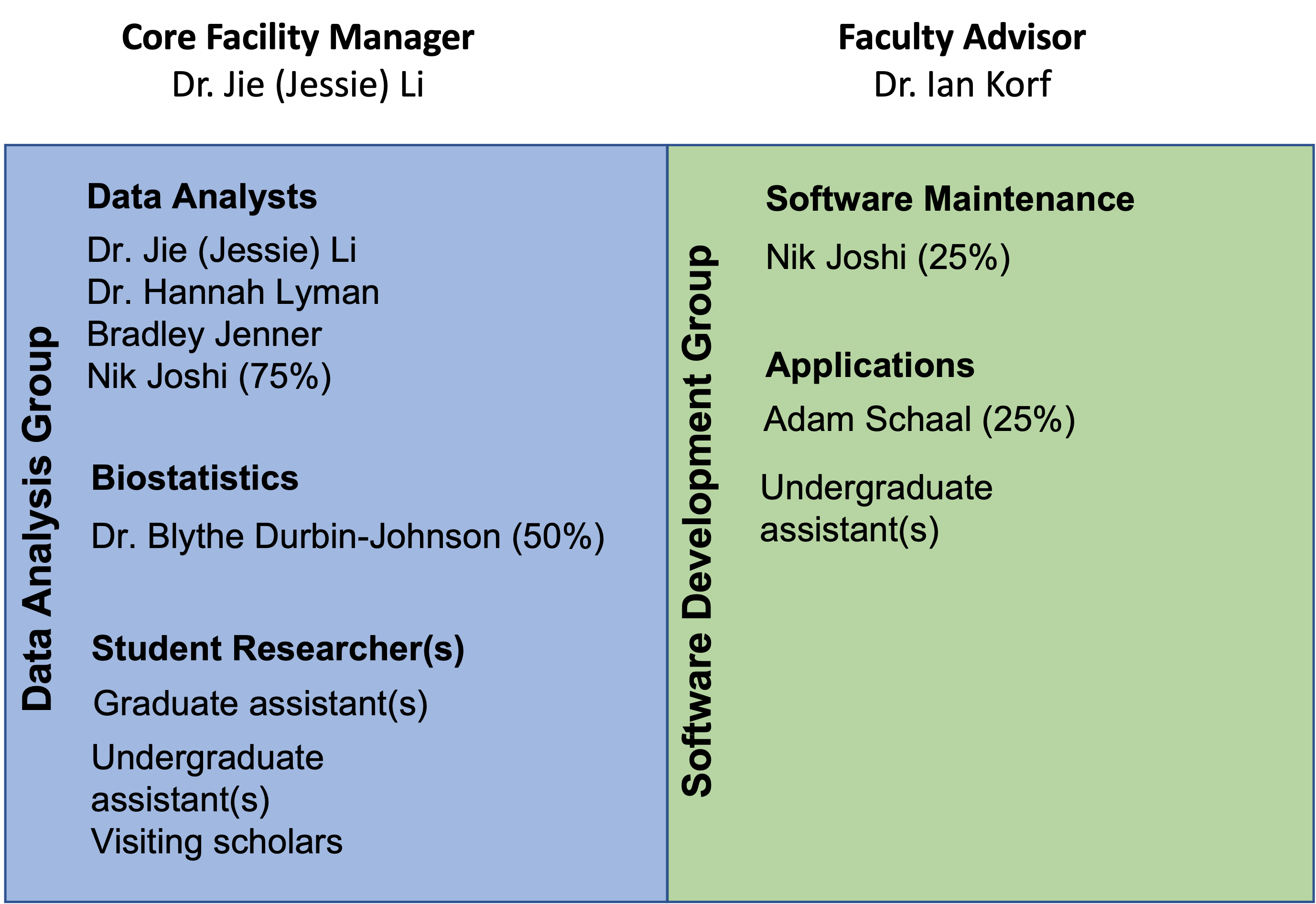
Welcome Figure2 This workshop will instruct participants on how to design a single cell rna seq experiment, and how to efficiently manage and analyze the data starting from count matrices. We start from the output of the cell ranger preprocessing software. this is an open source software suite that allows to pre process the fastq files generated by the sequencing platform and perform alignment and quantification. In this workshop we have walked through an end to end workflow for the analysis of single cell rna seq data. for each step, code was provided along with in depth information on the background and theory. What are umis? unique molecular identifiers give (almost) exact molecule counts in sequencing experiments. they reduce the amplification noise by allowing (almost) complete de duplication of sequenced fragments.

Single Cell Rna Sequencing Technologies And Bioinformatics 59 Off In this workshop we have walked through an end to end workflow for the analysis of single cell rna seq data. for each step, code was provided along with in depth information on the background and theory. What are umis? unique molecular identifiers give (almost) exact molecule counts in sequencing experiments. they reduce the amplification noise by allowing (almost) complete de duplication of sequenced fragments. Use seurat and associated tools to perform analysis of single cell expression data, including data filtering, qc, integration, clustering, and marker identification. To get the umis’ counts, we can first group reads by cell barcode before aligning cdna reads and counting unique molecules per cell per gene using the umis. figure 1.4: grouping barcodes to assign reads to cells (modified from david tse et.al). A well planned pilot experiment is essential for evaluating sample preparation and for understanding the required number of cells. you need your cells to be highly viable (>90 95%), have no clumps and no debris. cell membrane integrity is a must! free floating rna will make analysis more challenging. This workshop will instruct participants on how to design a single cell rna seq experiment, and how to efficiently manage and analyze the data starting from count matrices.
Comments are closed.