Ensemble Models And Use Of Feature Importance In Tree Based Models
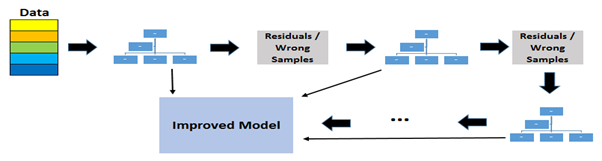
Ensemble Models And Use Of Feature Importance In Tree Based Models Ensembl is a genome browser for vertebrate genomes that supports research in comparative genomics, evolution, sequence variation and transcriptional regulation. ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. ensembl tools include blast, blat, biomart and the variant effect predictor (vep) for all supported species. ensembl release. We would like to show you a description here but the site won’t allow us.

Ensemble Models And Use Of Feature Importance In Tree Based Models About ensembl variation the ensembl variation database stores areas of the genome that differ between individual genomes ("variants") and, where available, associated disease and phenotype information. there are different types of variants for human, including some with restricted access: single nucleotide polymorphisms (snps) short nucleotide insertions and or deletions longer variants. Genome assembly: grch38.p14 (gca 000001405.29) more information and statistics download dna sequence (fasta) convert your data to grch38 coordinates display your data in ensembl other assemblies. Browse a genome the ensembl project produces genome databases for vertebrates and other eukaryotic species, and makes this information freely available online. Blast stands for basic local alignment search tool. the emphasis of this tool is to find regions of sequence similarity, which will yield functional and evolutionary clues about the structure and function of your sequence.
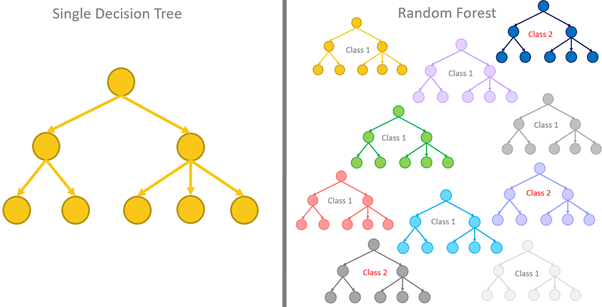
Ensemble Models And Use Of Feature Importance In Tree Based Models Browse a genome the ensembl project produces genome databases for vertebrates and other eukaryotic species, and makes this information freely available online. Blast stands for basic local alignment search tool. the emphasis of this tool is to find regions of sequence similarity, which will yield functional and evolutionary clues about the structure and function of your sequence. About archive ensembl the main ensembl site ( ensembl.org) and the mirror sites are updated with the latest data approximately every three months. we maintain the ensembl archive sites so that there are stable links to data from a particular release. as of december 2016 these will be available for five years, together with the following longer term archives: annotation on the human ncbi36. Use the vep online to analyse your variants through a simple point and click interface. the web interface allows you to access the key features of the vep without using the command line. interactively filter your results to find the data you want. download your results in multiple data formats, easily share your results with others, and integrate your variation data with the powerful ensembl. Ensembl release 114 may 2025 © embl ebipermanent link view in archive site. Homology types using the gene trees, we can infer the following pairwise relationships. orthologues homologues which diverged by a speciation event. there are four types of orthologues: 1 to 1 orthologues (ortholog one2one) 1 to many orthologues (ortholog one2many) many to many orthologues (ortholog many2many) between species paralogues – only as exceptions genes in different species and.
Ensemble Models And Use Of Feature Importance In Tree Based Models About archive ensembl the main ensembl site ( ensembl.org) and the mirror sites are updated with the latest data approximately every three months. we maintain the ensembl archive sites so that there are stable links to data from a particular release. as of december 2016 these will be available for five years, together with the following longer term archives: annotation on the human ncbi36. Use the vep online to analyse your variants through a simple point and click interface. the web interface allows you to access the key features of the vep without using the command line. interactively filter your results to find the data you want. download your results in multiple data formats, easily share your results with others, and integrate your variation data with the powerful ensembl. Ensembl release 114 may 2025 © embl ebipermanent link view in archive site. Homology types using the gene trees, we can infer the following pairwise relationships. orthologues homologues which diverged by a speciation event. there are four types of orthologues: 1 to 1 orthologues (ortholog one2one) 1 to many orthologues (ortholog one2many) many to many orthologues (ortholog many2many) between species paralogues – only as exceptions genes in different species and.

Ensemble Models And Use Of Feature Importance In Tree Based Models Ensembl release 114 may 2025 © embl ebipermanent link view in archive site. Homology types using the gene trees, we can infer the following pairwise relationships. orthologues homologues which diverged by a speciation event. there are four types of orthologues: 1 to 1 orthologues (ortholog one2one) 1 to many orthologues (ortholog one2many) many to many orthologues (ortholog many2many) between species paralogues – only as exceptions genes in different species and.

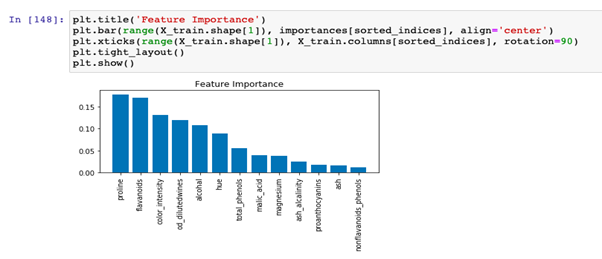
Feature Importance Based On All Regression Tree Models Download
Comments are closed.