Bcftools Tutorial On How To Count The Number Of Variants Per Chromosome In A Vcf File

Number Of Variants Per Chromosome Download Scientific Diagram Bcftools tutorial | bcftools view | count the number of variants per chromosome in a vcf file bioinformatics for beginners 3.42k subscribers subscribed. For each bcftools command, one would need to understand how that command treats multi allelic sites to ensure their results are accurate. run bcftools norm (or even better, vt norm vt decompose) before using bcftools.

Patient Variant Count And Distribution Per Chromosome Copy Number #select a particular genotype (0 1 or 1 1) from a vcf. in this case access sample accessed by index 8: this is great! a nice thing to possibly add: gist.github adefelicibus e668b2f03c157b3b272c871030c5d0b9. split vcf by sample: for sample in `bcftools view h $file | grep "^#chrom" | cut f10 `; do. In this tutorial we will first do variant calling using bcftools, and then we will explore the resulting files a bit. How to count variants per chromosome per sample in a cf file using bcftools | patreon exclusive bioinformatics coach 20.5k subscribers subscribed. I would like to count the number of snps and the number of indels per sample. the vcf files correspond to 30 samples, from which i am interested in 11, that i have listed in my samples list.txt. my code: n indels=$(bcftools view s $sample name recal indels.vcf | grep v "^#" | wc l).

Counts Of Variants Per Chromosome Download Table How to count variants per chromosome per sample in a cf file using bcftools | patreon exclusive bioinformatics coach 20.5k subscribers subscribed. I would like to count the number of snps and the number of indels per sample. the vcf files correspond to 30 samples, from which i am interested in 11, that i have listed in my samples list.txt. my code: n indels=$(bcftools view s $sample name recal indels.vcf | grep v "^#" | wc l). How to quickly count the number of genetic variants in a vcf file? you can also use bcftools to quickly count how many variants or rows there are in your file using the following combination of bcftools and the wc (word count) command:. Discover how to count variants per chromosome, split vcf files into snps and indels, and subset files based on sample ids. gain practical knowledge to enhance your bioinformatics toolkit and improve your ability to analyze genetic variation data effectively. For this tutorial, we will use bcftools which is designed by the same team behind samtools they are part of the same pipeline. bcftools is itself a comprehensive pipeline and produces a variant call format (vcf) that is used in many downstream analyses. Bcftools is a set of utilities that manipulate variant calls in the variant call format (vcf) and its binary counterpart bcf. all commands work transparently with both vcfs and bcfs, both uncompressed and bgzf compressed.
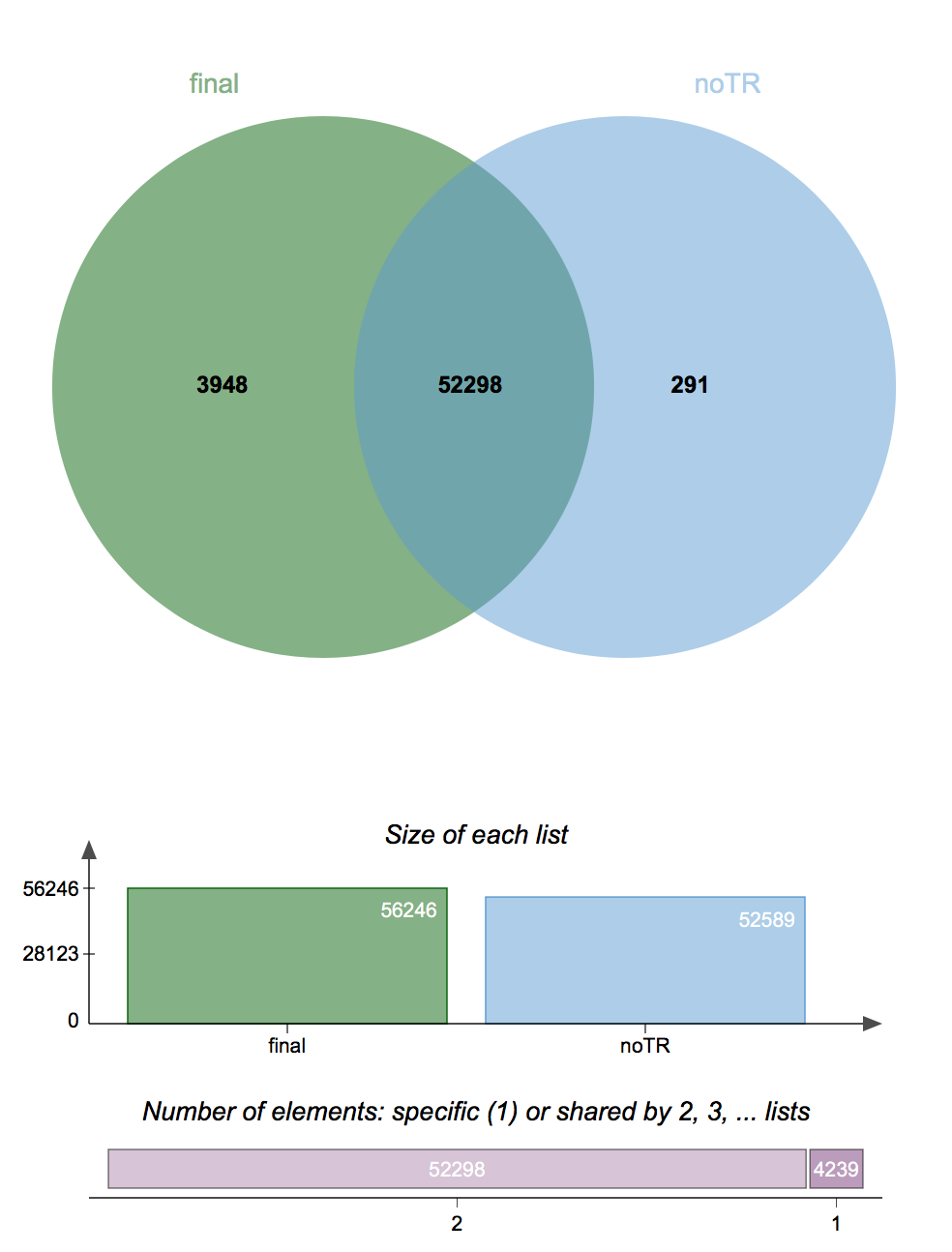
How To Find Common Variants In Multiple Vcf Files Blog How to quickly count the number of genetic variants in a vcf file? you can also use bcftools to quickly count how many variants or rows there are in your file using the following combination of bcftools and the wc (word count) command:. Discover how to count variants per chromosome, split vcf files into snps and indels, and subset files based on sample ids. gain practical knowledge to enhance your bioinformatics toolkit and improve your ability to analyze genetic variation data effectively. For this tutorial, we will use bcftools which is designed by the same team behind samtools they are part of the same pipeline. bcftools is itself a comprehensive pipeline and produces a variant call format (vcf) that is used in many downstream analyses. Bcftools is a set of utilities that manipulate variant calls in the variant call format (vcf) and its binary counterpart bcf. all commands work transparently with both vcfs and bcfs, both uncompressed and bgzf compressed.

Variant Annotation Speed And Ram Benchmarks On Variants From Genome For this tutorial, we will use bcftools which is designed by the same team behind samtools they are part of the same pipeline. bcftools is itself a comprehensive pipeline and produces a variant call format (vcf) that is used in many downstream analyses. Bcftools is a set of utilities that manipulate variant calls in the variant call format (vcf) and its binary counterpart bcf. all commands work transparently with both vcfs and bcfs, both uncompressed and bgzf compressed.
Comments are closed.